Note
Go to the end to download the full example code or to run this example in your browser via Binder
Train Models¶
Load PiML
from piml import Experiment
exp = Experiment()
exp.data_loader(data='CaliforniaHousing_raw')
exp.data_prepare(target='MedHouseVal', task_type='regression', random_state=0)
MedInc HouseAge AveRooms AveBedrms Population AveOccup Latitude \
0 8.3252 41.0 6.984127 1.023810 322.0 2.555556 37.88
1 8.3014 21.0 6.238137 0.971880 2401.0 2.109842 37.86
2 7.2574 52.0 8.288136 1.073446 496.0 2.802260 37.85
3 5.6431 52.0 5.817352 1.073059 558.0 2.547945 37.85
4 3.8462 52.0 6.281853 1.081081 565.0 2.181467 37.85
... ... ... ... ... ... ... ...
20635 1.5603 25.0 5.045455 1.133333 845.0 2.560606 39.48
20636 2.5568 18.0 6.114035 1.315789 356.0 3.122807 39.49
20637 1.7000 17.0 5.205543 1.120092 1007.0 2.325635 39.43
20638 1.8672 18.0 5.329513 1.171920 741.0 2.123209 39.43
20639 2.3886 16.0 5.254717 1.162264 1387.0 2.616981 39.37
Longitude MedHouseVal
0 -122.23 4.526
1 -122.22 3.585
2 -122.24 3.521
3 -122.25 3.413
4 -122.25 3.422
... ... ...
20635 -121.09 0.781
20636 -121.21 0.771
20637 -121.22 0.923
20638 -121.32 0.847
20639 -121.24 0.894
[20640 rows x 9 columns]
Config Value
0 Excluded columns []
1 Target variable MedHouseVal
2 Sample weight None
3 Task type regression
4 Split method random
5 Test ratio 0.2
6 Random state 0
Train and Register Models using piml
from lightgbm import LGBMRegressor
lgbm2 = LGBMRegressor(max_depth=2)
exp.model_train(lgbm2, name='LGBM_2')
Save Fitted Models
exp.model_save("LGBM_2", "CH_LGBM_2.pkl")
Load model from file system, if not specified, the default train and test data will be used.
pipeline = exp.make_pipeline(model='CH_LGBM_2.pkl')
exp.register(pipeline, "LGBM_2_load")
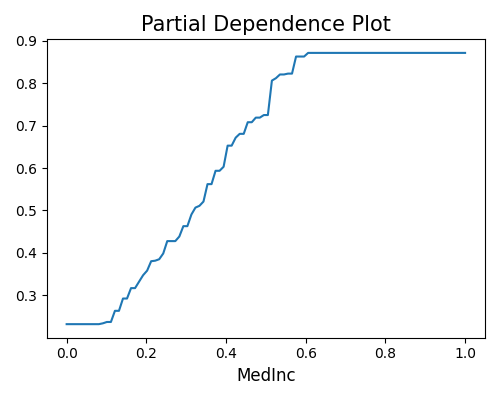
Run post-hoc explanation using PDP.
exp.model_explain(model="LGBM_2_load", show="pdp", uni_feature="MedInc", figsize=(5, 4))
Total running time of the script: ( 0 minutes 38.674 seconds)
Estimated memory usage: 19 MB
