Note
Go to the end to download the full example code or to run this example in your browser via Binder
Register H2O Models¶
Assume we have fitted a H2O model outside PiML workflow
For demonstration, we fit a model using H2O
import h2o
h2o.no_progress()
h2o.init(verbose=False)
import numpy as np
import pandas as pd
from sklearn.datasets import fetch_california_housing
from h2o.estimators import H2OGradientBoostingEstimator
data = fetch_california_housing()
feature_names = data.feature_names
target_name = data.target_names[0]
h2o_data = h2o.H2OFrame(pd.DataFrame(np.hstack([data.data, data.target.reshape(-1, 1)]),
columns=feature_names + [target_name])
)
h2o_data_train, h2o_data_test = h2o_data.split_frame(ratios=[0.8], seed=2023)
gbm_model = H2OGradientBoostingEstimator()
gbm_model.train(feature_names, target_name, training_frame=h2o_data_train)
# Save the model to file system
mojo_file_path = gbm_model.save_mojo(path="./")
Then, we can test this model using PiML
from piml import Experiment
exp = Experiment(highcode_only=True)
Load the MOJO model into memory and register it into PiML
imported_model = h2o.import_mojo(mojo_file_path)
pipeline = exp.make_pipeline(model=imported_model,
task_type="regression",
train_x=h2o_data_train[feature_names].as_data_frame().values,
train_y=h2o_data_train[target_name].as_data_frame().values.ravel(),
test_x=h2o_data_test[feature_names].as_data_frame().values,
test_y=h2o_data_test[target_name].as_data_frame().values.ravel(),
feature_names=feature_names,
target_name=target_name)
exp.register(pipeline, "H2O-GBM")
Check model performance
exp.model_diagnose(model="H2O-GBM", show="accuracy_table")
MSE MAE R2
Train 0.2423 0.3396 0.8180
Test 0.2377 0.3429 0.8216
Gap -0.0046 0.0033 0.0037
Explain using post-hoc explanation tools
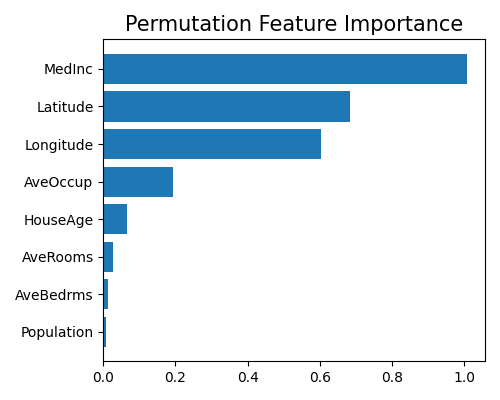
exp.model_explain(model="H2O-GBM", show="pfi", figsize=(5, 4))
Run validataion tests
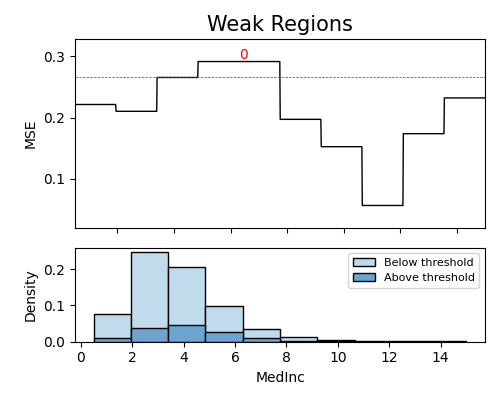
exp.model_diagnose(model="H2O-GBM", show="weakspot", slice_features=["MedInc"], figsize=(5, 4))
Total running time of the script: ( 5 minutes 55.568 seconds)
Estimated memory usage: 34 MB
