Note
Go to the end to download the full example code or to run this example in your browser via Binder
Register Arbitrary Models¶
If the model is neither sklearn style nor H2O style, we can still register it into PiML.
For demonstration, we simulate a simple binary classification dataset and fit a GLM using statsmodels.
import numpy as np
import statsmodels.api as sm
x = np.random.uniform(-1, 1, size=(1000, 2))
y = (np.sum(x, axis=1) + np.random.normal(0, 0.1, size=(1000,))) > 0.0
glm_binom = sm.GLM(y, x, family=sm.families.Binomial())
glm_results = glm_binom.fit()
Next, we define the wrapper functions of predict and predict_proba.
def predict_proba_func(X):
proba = glm_binom.predict(glm_results.params, exog=X)
return np.vstack([1 - proba, proba]).T
def predict_func(X):
proba = glm_binom.predict(glm_results.params, exog=X)
return proba > 0.5
Register the fitted model into PiML (please make sure the datasets of different pipelines are the same)
from piml import Experiment
exp = Experiment(highcode_only=True)
pipeline = exp.make_pipeline(predict_func=predict_func,
predict_proba_func=predict_proba_func,
task_type="classification",
train_x=x[:800],
train_y=y[:800],
test_x=x[800:],
test_y=y[800:],
feature_names=["X0", "X1"],
target_name="Y")
exp.register(pipeline, "Statsmodels-GLM")
Check model performance
exp.model_diagnose(model="Statsmodels-GLM", show="accuracy_table")
ACC AUC F1 LogLoss Brier
Train 0.9700 0.9971 0.9704 0.0690 0.0216
Test 0.9750 0.9961 0.9730 0.0791 0.0220
Gap 0.0050 -0.0010 0.0026 0.0102 0.0003
Explain using post-hoc explanation tools
exp.model_explain(model="Statsmodels-GLM", show="pfi", figsize=(5, 4))
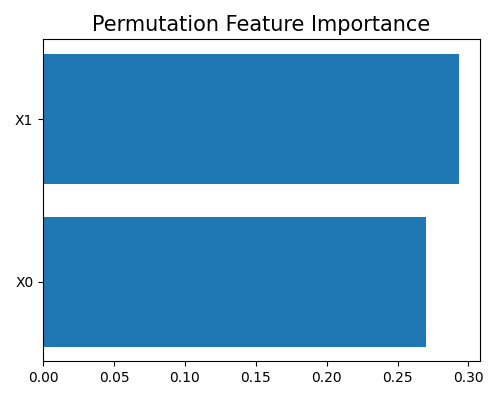
Explain using post-hoc explanation tools
exp.model_explain(model="Statsmodels-GLM", show="ale", uni_feature="X0", figsize=(5, 4))
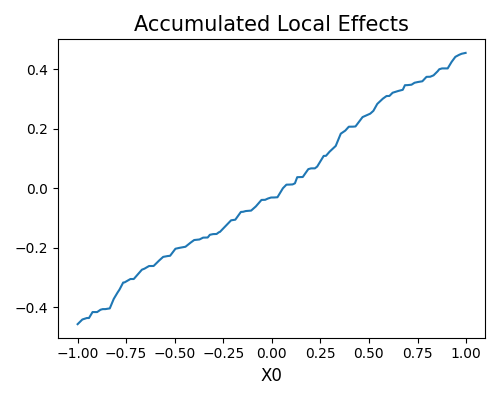
Run validataion tests
exp.model_explain(model="Statsmodels-GLM", show="pdp", uni_feature="X0", figsize=(5, 4))
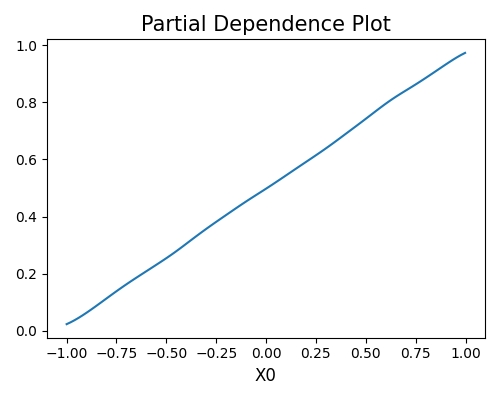
Total running time of the script: ( 0 minutes 32.525 seconds)
Estimated memory usage: 24 MB
