Note
Go to the end to download the full example code or to run this example in your browser via Binder
XGB-1 Regression (California Housing)¶
Experiment initialization and data preparation
from piml import Experiment
from piml.models import XGB1Regressor
exp = Experiment()
exp.data_loader(data="CaliforniaHousing_trim2", silent=True)
exp.data_prepare(target="MedHouseVal", task_type="regression", silent=True)
Train model
exp.model_train(model=XGB1Regressor(n_estimators=500, max_bin=20, min_bin_size=0.01), name="XGB1")
Train model with monotonicity constrained on MedInc
exp.model_train(model=XGB1Regressor(n_estimators=500, max_bin=20, min_bin_size=0.01,
mono_increasing_list=("MedInc", )),
name="Mono-XGB1")
Evaluate predictive performance of XGB1
exp.model_diagnose(model="XGB1", show='accuracy_table')
MSE MAE R2
Train 0.0131 0.0816 0.7704
Test 0.0136 0.0822 0.7546
Gap 0.0005 0.0006 -0.0159
Evaluate predictive performance of Mono-XGB1
exp.model_diagnose(model="Mono-XGB1", show='accuracy_table')
MSE MAE R2
Train 0.0134 0.0831 0.7649
Test 0.0139 0.0837 0.7490
Gap 0.0005 0.0006 -0.0159
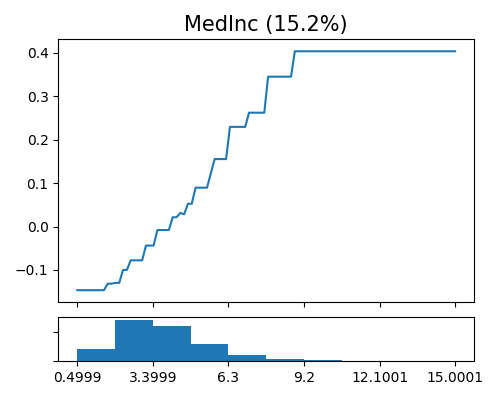
Global effect plot of XGB1 on MedInc
exp.model_interpret(model="XGB1", show="global_effect_plot", uni_feature="MedInc",
original_scale=True, figsize=(5, 4))
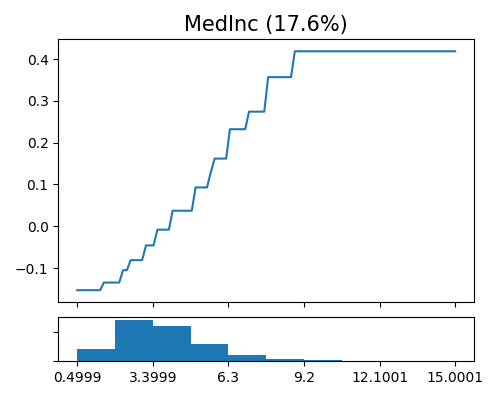
Global effect plot of Mono-XGB1 on MedInc
exp.model_interpret(model="Mono-XGB1", show="global_effect_plot", uni_feature="MedInc",
original_scale=True, figsize=(5, 4))
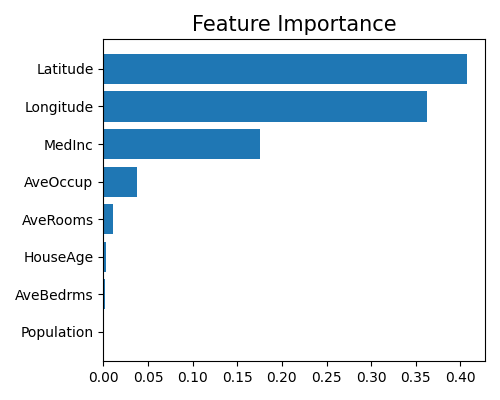
Feature importance of Mono-XGB1
exp.model_interpret(model="Mono-XGB1", show="global_fi", figsize=(5, 4))
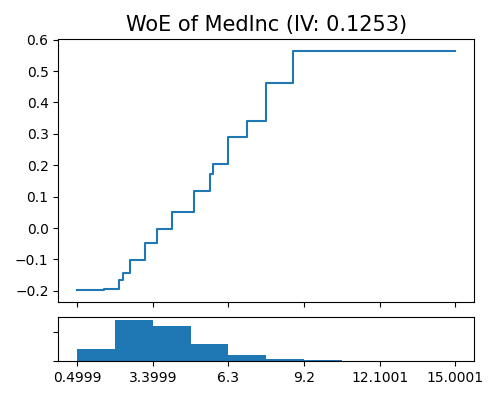
Weight of evidence plot of Mono-XGB1 on MedInc
exp.model_interpret(model="Mono-XGB1", show="xgb1_woe", uni_feature="MedInc", original_scale=True, figsize=(5, 4))
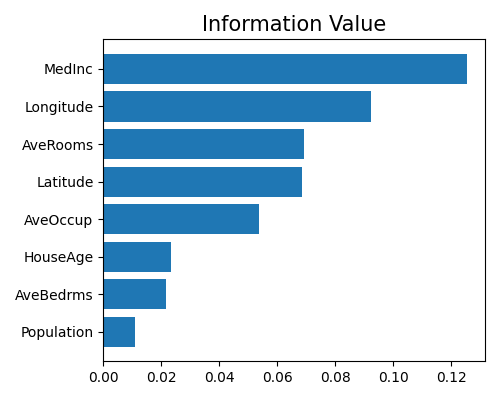
Information value plot of Mono-XGB1
exp.model_interpret(model="Mono-XGB1", show="xgb1_iv", figsize=(5, 4))
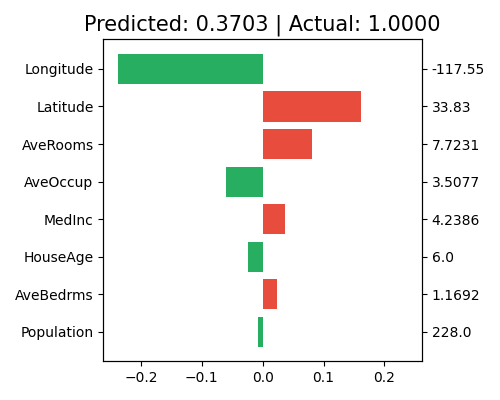
Local interpretation of Mono-XGB1
exp.model_interpret(model="Mono-XGB1", show="local_fi", sample_id=0, original_scale=True, figsize=(5, 4))
Total running time of the script: (0 minutes 8.159 seconds)
