6.8. Segmented¶
Segmented test is a separate model diagnostic module that is designed to diagnose the performance of a model in different segments. It is a powerful tool that can help users identify the weak regions of a model and provide insights into the root causes of the underperformance. In PiML, it can be triggered by the exp.segmented_diagnose function.
6.8.1. Methodology¶
Once a model is fitted and registered in PiML, this test facilitates a detailed evaluation of model weaknesses. It begins by segmenting the data into distinct subsets based on a selected feature of interest, followed by individual analyses for each subset. Notably, three segmentation methods are available:
‘uniform’: Segmentation based on equal intervals.
‘quantile’: Segmentation based on equal quantiles.
‘auto’: Automatic segmentation of feature segments determined by a surrogate model. The process involves fitting an XGB1 model between covariates and the residual, extracting split points from the XGB1 model, and using these points to segment the data. It’s important to note that some features may have no split points.
6.8.2. Usage¶
The exp.segmented_diagnose function has the following arguments.
show: The available test keywords include‘segment_table’: the accuracy of each segment. If
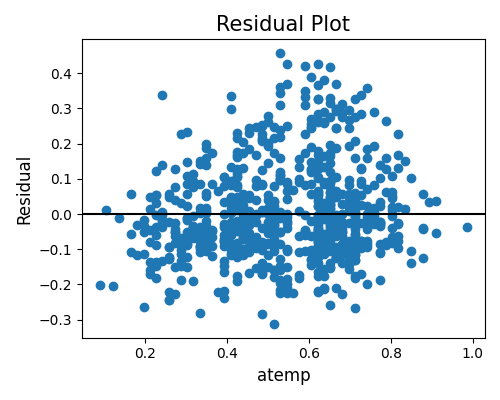
segment_feature= None, the results for all features will be output.‘accuracy_residual’: the residual plot of the specified segment and feature of interest.
‘accuracy_table’: the accuracy results of the specified segment.
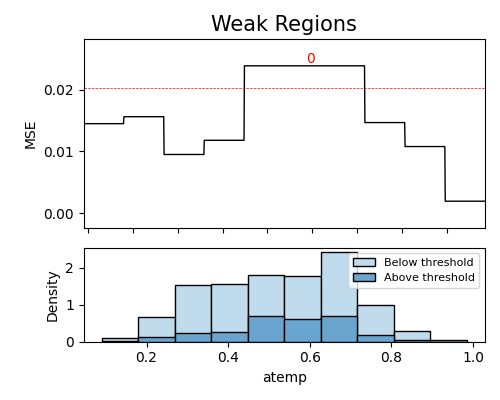
‘weakspot’: the weakspot test of the specified segment.
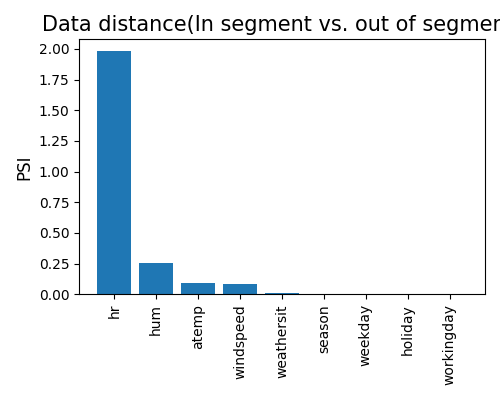
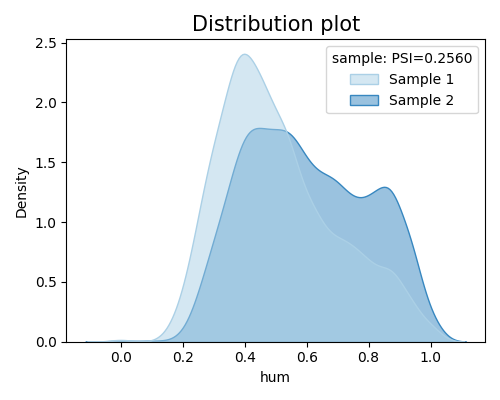
‘distribution_shift’: the density plot comparison between the specified segment and its complement.
segment_method: The segmentation method, including “uniform”, “quantile”, and “auto”.segment_bins: The number of bins, used whensegment_methodis “uniform” or “quantile”.segment_feature: Feature filter for bucketing, only works whenshow= “segment_table”.segment_id: The ID of the segment to diagnose.metric: The performance metric for evaluation, which includes “MSE”, “MAE”, and “R2” for regression tasks, and “ACC”, “AUC”, ‘LogLoss’, ‘Brier’ and “F1” for classification tasks. The default metric is “MSE” for regression and “ACC” for classification.
6.8.2.1. Segments summary¶
In the segmented diagnose module, you can start by obtaining a summary of the segments, by setting the parameter show to ‘segment_table’. This table provides a summary of all segments and helps users quickly identify any regions of interest. Additionally, if you define the segment_feature parameter, the table will be filtered to only show the feature-related results of the segments.
result = exp.segmented_diagnose(model='XGB2', show='segment_table',
segment_method='uniform', segment_bins=5, return_data=True)
result.data
The table displayed above presents the top 10 weakest segments. It consists of the following columns:
Segment ID: The ID of each segment.
Feature: The segmented feature.
Segment: The segment range if it is a numerical feature, segment value if it is a categorical feature.
Size: The number of samples contained within each segment.
metric: The metric value corresponding to each segment.
6.8.2.2. Accuracy Table¶
Once you have specified a specific segment_id, you can further analyze the performance of that segment. By setting show to ‘accuracy_table’, an accuracy table will be presented. This table provides information and insights regarding the accuracy of the segment you have chosen. It can provide valuable metrics and metrics-based analysis to help you understand the performance of that segment. For more details about the metrics, you can see: test_accuracy.
result = exp.segmented_diagnose(model='XGB2', show='accuracy_table', segment_id=0, return_data=True)
result.data.head(10)